DNA...From Blueprint to Brick
In 2005, Caltech researcher Paul W. K. Rothemund made a smiley face. In fact, he made about 50 billion of them. Other than the sheer amount, what was remarkable about the accomplishment was what he made his smiley faces from -- DNA.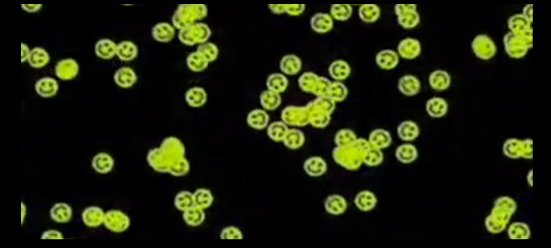 Rothemund's tour de force lab accomplishment was one of the most highly visible milestones in the emergence of a new scientific community. Rather than seeing DNA as primarily an information containing molecule - a blueprint - DNA nanotechnologists treat the iconic molecule as something to build with - a brick.For much of the late 20th century, scientists, writers, and the general public imagined DNA as information. It was code in the form of a chemical, a molecule that directed our development and determined our destiny. This discourse served to organize, guide, and inform the research agenda of scientists for decades.Starting in the late 1970s, an interdisciplinary group of chemists, crystallographers, molecular biologists and computer scientists began to reconceptualize what DNA was and what people might be able to do with it. The main person - for years, really the only person - at the vanguard of this effort was Nadrian "Ned" Seeman. In the late 1970s, Seeman, a biochemist whose main field of expertise was crystallography, was an assistant professor languishing in the biology department at SUNY-Albany.
Rothemund's tour de force lab accomplishment was one of the most highly visible milestones in the emergence of a new scientific community. Rather than seeing DNA as primarily an information containing molecule - a blueprint - DNA nanotechnologists treat the iconic molecule as something to build with - a brick.For much of the late 20th century, scientists, writers, and the general public imagined DNA as information. It was code in the form of a chemical, a molecule that directed our development and determined our destiny. This discourse served to organize, guide, and inform the research agenda of scientists for decades.Starting in the late 1970s, an interdisciplinary group of chemists, crystallographers, molecular biologists and computer scientists began to reconceptualize what DNA was and what people might be able to do with it. The main person - for years, really the only person - at the vanguard of this effort was Nadrian "Ned" Seeman. In the late 1970s, Seeman, a biochemist whose main field of expertise was crystallography, was an assistant professor languishing in the biology department at SUNY-Albany. Seeman worked with complex organic molecules. These are notoriously difficult to crystallize. As a result, Seeman, about to come up for tenure, faced a dilemma captured nicely in the image below. Basically, "no crystals, no crystallography, no crystallographer."
Seeman worked with complex organic molecules. These are notoriously difficult to crystallize. As a result, Seeman, about to come up for tenure, faced a dilemma captured nicely in the image below. Basically, "no crystals, no crystallography, no crystallographer." At the same time, Seeman was thinking about what it might be possible to build with DNA. Why DNA? First of all, it's well studied - what historians of science call a model system. DNA's structure is predictable, made of four different types of nucleotide subunits—adenine, cytosine, guanine, and thymine. The exact sequence of an organism’s DNA is determined by what scientists call complementary base pairing: adenine always pairs with thymine; guanine connects with cytosine.
At the same time, Seeman was thinking about what it might be possible to build with DNA. Why DNA? First of all, it's well studied - what historians of science call a model system. DNA's structure is predictable, made of four different types of nucleotide subunits—adenine, cytosine, guanine, and thymine. The exact sequence of an organism’s DNA is determined by what scientists call complementary base pairing: adenine always pairs with thymine; guanine connects with cytosine.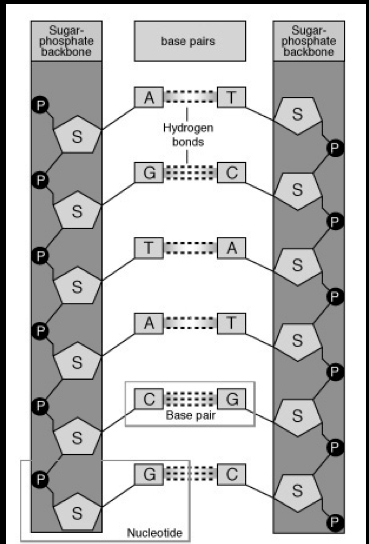 This predictability allows scientists to synthesize strands of artificial DNA—a technique perfected and automated in the 1980s—which, when properly treated in the lab, can link up to form a desired structure.Seeman was thinking about a particular form of DNA that occurs during a process known as genetic recombination. This involves the breaking and rejoining of two homologous DNA double helices as shown below. If the two DNA molecules have regions of similar nucleotide sequences, they can “cross over” and form a novel nucleotide sequence. A crucial intermediate stage in this recombination of DNA is a structure known as a Holliday junction (named after the British molecular biologist who first proposed it in 1964).
This predictability allows scientists to synthesize strands of artificial DNA—a technique perfected and automated in the 1980s—which, when properly treated in the lab, can link up to form a desired structure.Seeman was thinking about a particular form of DNA that occurs during a process known as genetic recombination. This involves the breaking and rejoining of two homologous DNA double helices as shown below. If the two DNA molecules have regions of similar nucleotide sequences, they can “cross over” and form a novel nucleotide sequence. A crucial intermediate stage in this recombination of DNA is a structure known as a Holliday junction (named after the British molecular biologist who first proposed it in 1964).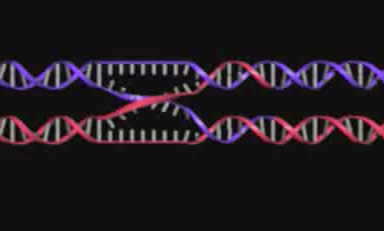 In 1979, Seeman started doing computer modeling of these Holliday junctions. His goal was to try to better understand their motion during recombination. Seeman soon recognized that in principle one could – using synthetic DNA – create junctions that didn’t move.To do this, Seeman would have to take advantage of another feature of DNA. This is called a “sticky end.” These occur when one strand of the double helix extends several base pairs beyond the other strand. This presence of sticky ends meant, Seeman reasoned, that one could – using junctions as well as sticky ended cohesion – build DNA structures that formed lattices and networks.In Seeman's telling, his epiphany came while sitting in a bar in Albany. Crystallographers, not surprisingly, are quite familiar, even fond, of works by M.C. Escher given the Dutch artist's focus on periodicity, symmetry, and the overall representation of objects in space. Seeman recalled Escher's 1955 painting Depth:
In 1979, Seeman started doing computer modeling of these Holliday junctions. His goal was to try to better understand their motion during recombination. Seeman soon recognized that in principle one could – using synthetic DNA – create junctions that didn’t move.To do this, Seeman would have to take advantage of another feature of DNA. This is called a “sticky end.” These occur when one strand of the double helix extends several base pairs beyond the other strand. This presence of sticky ends meant, Seeman reasoned, that one could – using junctions as well as sticky ended cohesion – build DNA structures that formed lattices and networks.In Seeman's telling, his epiphany came while sitting in a bar in Albany. Crystallographers, not surprisingly, are quite familiar, even fond, of works by M.C. Escher given the Dutch artist's focus on periodicity, symmetry, and the overall representation of objects in space. Seeman recalled Escher's 1955 painting Depth: Seeman realized that the fish in the Escher picture were just like a 6-arm junction arrayed in periodic fashion. Wondering if he might be able to make a similar structure from branched DNA molecules, Seeman proposed his ideas in an article that came out in the Journal of Theoretical Biology in early 1982.
Seeman realized that the fish in the Escher picture were just like a 6-arm junction arrayed in periodic fashion. Wondering if he might be able to make a similar structure from branched DNA molecules, Seeman proposed his ideas in an article that came out in the Journal of Theoretical Biology in early 1982. It’s important to note where Seeman’s paper – now cited nearly 900 times – appeared. It was in a journal of theoretical biology. Seeman was noting that it was possible to make these DNA lattices…but he had yet to demonstrate this could actually be done in the lab.Doing this required overcoming one key obstacle - getting the right amount and right sequence of DNA. In the early 1980s, one could buy synthetic DNA but it was very expensive. A strand of DNA with the desired order of base pairs, maybe only 10-12 nucleotides, might cost around $6000.
It’s important to note where Seeman’s paper – now cited nearly 900 times – appeared. It was in a journal of theoretical biology. Seeman was noting that it was possible to make these DNA lattices…but he had yet to demonstrate this could actually be done in the lab.Doing this required overcoming one key obstacle - getting the right amount and right sequence of DNA. In the early 1980s, one could buy synthetic DNA but it was very expensive. A strand of DNA with the desired order of base pairs, maybe only 10-12 nucleotides, might cost around $6000. So, Seeman opted for the cheaper but more time consuming option. Over the next several years, he learned the arcane craft of making DNA with custom sequences of nucleotide bases. A DNA synthesizer paid for by the National Institutes of Health became his lab’s most important piece of equipment.In late 1983, Seeman finally published a paper in Nature that gave experimental proof of the theoretical idea he had published 18 months earlier. Working with Neville Kallenbach, then at University of Pennsylvania, they demonstrated that it was possible to construct an immobile DNA junction. Kallenbach left Penn for New York University and in 1988, recruited Seeman to NYU’s Chemistry department.Two years later, working with his graduate student Junghuei Chen, he synthesized a DNA molecule that had ten strands. When combined with similar molecules, the result was a macromolecule with the “connectivity of a cube" – it was the first lab demonstration that one could make 3-D structures with DNA.
So, Seeman opted for the cheaper but more time consuming option. Over the next several years, he learned the arcane craft of making DNA with custom sequences of nucleotide bases. A DNA synthesizer paid for by the National Institutes of Health became his lab’s most important piece of equipment.In late 1983, Seeman finally published a paper in Nature that gave experimental proof of the theoretical idea he had published 18 months earlier. Working with Neville Kallenbach, then at University of Pennsylvania, they demonstrated that it was possible to construct an immobile DNA junction. Kallenbach left Penn for New York University and in 1988, recruited Seeman to NYU’s Chemistry department.Two years later, working with his graduate student Junghuei Chen, he synthesized a DNA molecule that had ten strands. When combined with similar molecules, the result was a macromolecule with the “connectivity of a cube" – it was the first lab demonstration that one could make 3-D structures with DNA. Seeman would later tell me that he always “regarded DNA as a four letter word” – referring to its four nucleotide bases and “not very interesting when it’s linear.” It was in the 3-D realm, building things with it that he found excitement.Today, DNA nanotechnology is one part of the growing field of synthetic biology. Today, more than 60 labs - including Paul Rothemund's at Caltech - are doing various forms of nanotechnology with DNA and RNA. To date, successes with DNA nanotechnology have included the construction of increasingly complex three-dimensional shapes, carrying out massively parallel computations, and building “DNA walkers” that can traverse a substrate and deliver “cargoes” of nanoscale particles.For a historian of science, what is fascinating about this evolving field is this new interpretation of DNA. DNA made the transition from genes to machines. In the end, it all comes back to a fundamental shift in how researchers saw DNA: from a code to a construction material.
Seeman would later tell me that he always “regarded DNA as a four letter word” – referring to its four nucleotide bases and “not very interesting when it’s linear.” It was in the 3-D realm, building things with it that he found excitement.Today, DNA nanotechnology is one part of the growing field of synthetic biology. Today, more than 60 labs - including Paul Rothemund's at Caltech - are doing various forms of nanotechnology with DNA and RNA. To date, successes with DNA nanotechnology have included the construction of increasingly complex three-dimensional shapes, carrying out massively parallel computations, and building “DNA walkers” that can traverse a substrate and deliver “cargoes” of nanoscale particles.For a historian of science, what is fascinating about this evolving field is this new interpretation of DNA. DNA made the transition from genes to machines. In the end, it all comes back to a fundamental shift in how researchers saw DNA: from a code to a construction material.